Note
Go to the end to download the full example code.
Gaussian Shells
Toy likelihood model for stress testing multiple-ellipsoid method.
The problem is:
\[\mathcal{L}(\theta) = \mathrm{circ}(\theta; c_1, r_1, w_1) +
\mathrm{circ}(\theta; c_2, r_2, w_2)\]
where
\[\mathrm{circ}(\theta; c, r, w) = \frac{1}{\sqrt{2 \pi w^2}}
\exp \left[ - \frac{(|\theta - c| - r)^2}{2 w^2} \right]\]
import math
import time
from collections import OrderedDict
import numpy as np
from numpy.random import RandomState
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import nestle
rstate = RandomState(0)
In the following block, we define the problem. We use r = 2 and w = 0.1, meaning that the gaussian is quite narrow compared to the size of the sphere.
r = 2.
w = 0.1
const = math.log(1. / math.sqrt(2. * math.pi * w**2))
def logcirc(theta, c):
d = np.sqrt(np.sum((theta - c)**2, axis=-1)) # |theta - c|
return const - (d - r)**2 / (2. * w**2)
def loglike(theta, c1, c2):
return np.logaddexp(logcirc(theta, c1), logcirc(theta, c2))
def prior_transform(x):
"""Defines a flat prior between -6 and 6 in all dimensions."""
return 12. * x - 6.
Visualize
It helps to visualize the surface in two dimensions. Here, we plot the likelihood evaluated on a fine grid and the sample points from nested sampling.
# likelihood surface in 2-d
xx, yy = np.meshgrid(np.linspace(-6., 6., 200),
np.linspace(-6., 6., 200))
c1 = np.array([-3.5, 0.])
c2 = np.array([3.5, 0.])
Z = np.exp(loglike(np.dstack((xx, yy)), c1, c2))
# nested sampling result
c1 = np.array([-3.5, 0.])
c2 = np.array([3.5, 0.])
f = lambda theta: loglike(theta, c1, c2)
res = nestle.sample(f, prior_transform, 2, method='multi', npoints=1000,
rstate=rstate)
fig = plt.figure(figsize=(14., 6.))
ax = fig.add_subplot(121, projection='3d')
ax.plot_surface(xx, yy, Z, rstride=1, cstride=1, linewidth=0, cmap='coolwarm')
ax.set_xlim(-6., 6.)
ax.set_ylim(-6., 6.)
ax.set_zlim(0., 4.)
ax.set_zlabel('L')
ax.set_title('Likelihood evaluated on fine grid')
ax = fig.add_subplot(122, projection='3d')
ax.scatter(res.samples[:,0], res.samples[:, 1], np.exp(res.logl),
marker='.', c=np.exp(res.logl), linewidths=(0.,), cmap='coolwarm')
ax.set_xlim(-6., 6.)
ax.set_ylim(-6., 6.)
ax.set_zlim(0., 4.)
ax.set_zlabel('L')
ax.set_title('Nested sampling points');
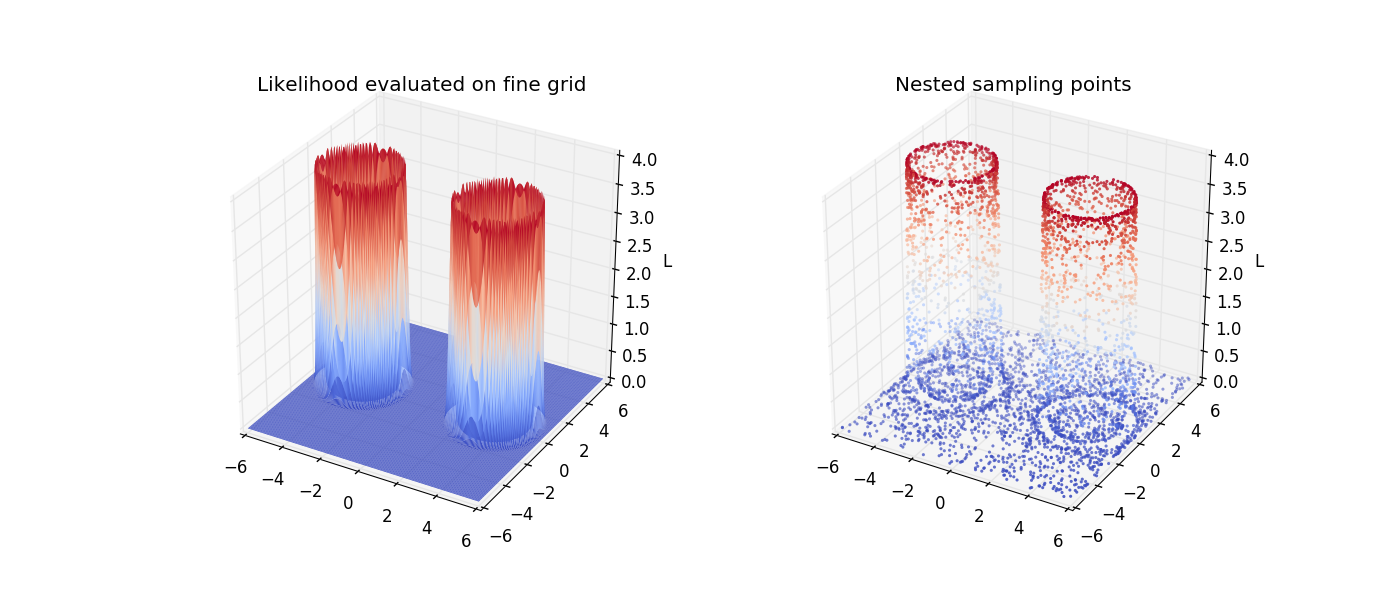
Text(0.5, 1.0, 'Nested sampling points')
Scaling with dimension
Here, we demonstrate how the algorithm scales with dimension and compare the total evidence to the analytic answer.
npoints = 1000
def run(ndim):
"""Convenience function for running in any dimension"""
c1 = np.zeros(ndim)
c1[0] = -3.5
c2 = np.zeros(ndim)
c2[0] = 3.5
f = lambda theta: loglike(theta, c1, c2)
return nestle.sample(f, prior_transform, ndim, method='multi',
npoints=npoints, rstate=rstate)
# Run over dimensions and save time for each run.
results = OrderedDict()
for ndim in [2, 5, 10, 20]:
t0 = time.time()
results[ndim] = run(ndim)
results[ndim].time = time.time() - t0
analytic_logz = {2: -1.75,
5: -5.67,
10: -14.59,
20: -36.09}
print("D analytic logz logzerr nlike eff(%) time")
for ndim, res in results.items():
eff = 100. * res.niter/(res.ncall - npoints)
print("{:2d} {:6.2f} {:6.2f} {:4.2f} {:6d} {:5.2f} {:6.2f}"
.format(ndim, analytic_logz[ndim], res.logz, res.logzerr,
res.ncall, eff, res.time))
D analytic logz logzerr nlike eff(%) time
2 -1.75 -1.79 0.05 11794 37.93 0.32
5 -5.67 -5.47 0.08 23061 35.23 0.66
10 -14.59 -14.31 0.12 48088 35.26 1.33
20 -36.09 -35.99 0.19 182116 21.13 6.99
Total running time of the script: (0 minutes 10.539 seconds)